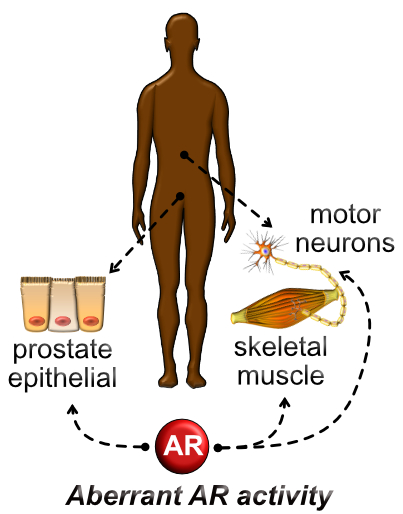
Androgen Receptor Signaling in AR-related Diseases
Androgen receptor belongs to a family of steroid hormone receptors. Androgen receptor is expressed in organs involved in human reproduction such as the prostate gland and ovaries, while also mediating actions in non-reproductive organs such as the brain, pancreas, liver, adipose tissue, and musculoskeletal system. We are studying how androgens regulate the cellular processes of differentiation, proliferation, and survival through the non-genomic and genomic actions of androgen receptor. Aberrant androgen receptor signaling contributes to multiple human diseases including prostate cancers, polycystic ovarian syndrome, spinal bulbar muscular atrophy, androgenetic alopecia, and androgen insensitivity syndrome. A major goal of the laboratory is to build quantitative models of androgen-mediated androgen receptor signal transduction that predict physiological and pathophysiological actions of androgen receptor at the molecular and cellular level. We are implementing cutting-edge proteomic and genomic technologies to interrogate androgen receptor function in cell-based models of human prostate cancer (e.g., AR mutations, AR isoform splice variants: AR-V7) and spinal bulbar muscular atrophy (e.g., polyglutamine expansion: AR-112Q).
Androgen-Regulated Protein Complexes and Modules
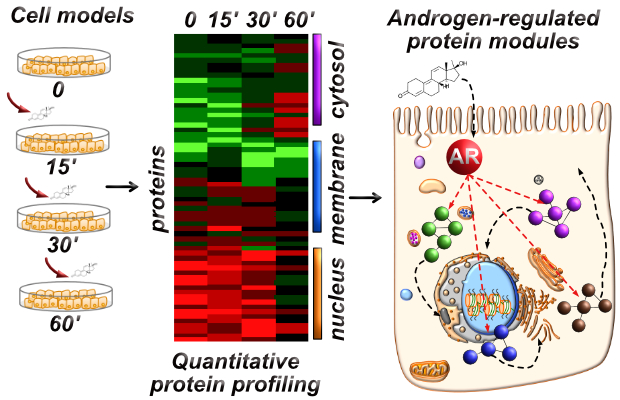
Androgens bind and activate androgen receptor to elicit numerous gene expression changes at the level of the transcriptome and proteome in hormone-responsive systems. We are focused on mapping acute, non-genomic protein abundance changes across the cytosolic, membrane, and nuclear compartments to understand how protein complexes and protein modules coordinate androgen-mediated androgen receptor activation. We are defining androgen-regulated proteomes in androgen-responsive systems using quantitative mass spectrometry to understand how this subproteome is coordinated under homeostatic conditions, and becomes corrupted to elicit aberrant androgen receptor functions in AR-related diseases.
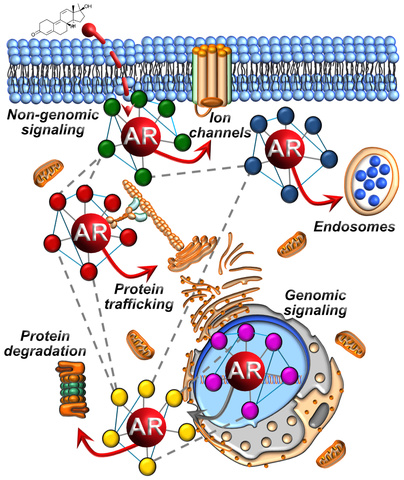
Genetic Analysis of the Androgen Receptor Interactome
Androgen receptor is a very sticky steroid hormone receptor that binds to more than 300 different proteins. These androgen receptor-interacting proteins, better known as the "androgen receptor interactome", encode critical regulators of homeostatic and pathologic androgen receptor signaling in AR-related diseases. Despite many years of research, how the androgen receptor interactome binds to androgen receptor to regulate androgen receptor function in space and time remains unresolved to date. My laboratory is defining spatial and temporal relationships between androgen receptor and the androgen receptor interactome utilizing proximity labeling coupled to quantitative mass spectrometry. A spatiotemporal understanding of the androgen receptor interactome will fill in the molecular gaps to discern how they individually, and collectively, regulate homeostatic and pathologic androgen receptor functions in cells, tissues, and organs. This new molecular information will be leveraged to define epistatic relationships, both positive and negative genetic interactions, across the androgen receptor interactome. We are probing genetic interactions across the androgen receptor interactome using genetically engineered CRISPRi and CRISPRa cell lines. CRISPR-mediated loss-of-function and gain-of-function experiments targeting the androgen receptor interactome will facilitate the molecular ordering of androgen-mediated AR signaling, both non-genomic and genomic, in hormone-responsive systems. More importantly, these results will provide a molecular framework to define and target synthetic lethal interactions in the androgen receptor interactome to attenuate aberrant androgen receptor activity underlying AR-related diseases.
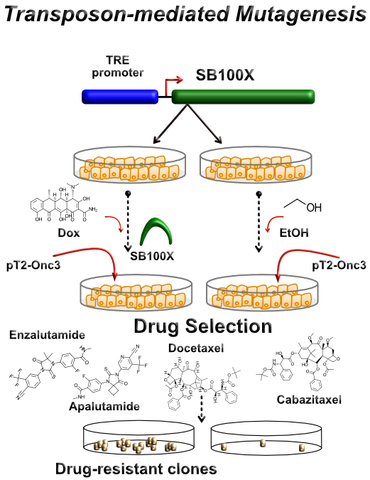
Probing Drug Resistance Pathways in Prostate Neoplasms
Androgen-deprivation therapy (ADT) coupled to taxane chemotherapy (e.g., docetaxel, paclitaxel) or androgen signaling inhibitors (ARSIs) (e.g., enzalutamide, abiraterone acetate) represent first-line therapies in the treatment of metastatic, hormone naïve prostate cancers. Unfortunately, these therapeutic regimens only prolong metastatic disease, as patients unavoidably develop incurable drug-resistant prostate cancer (CRPC). Identifying the mechanisms that confer drug-resistance upon CRPC is critical to identifying new druggable targets to cure CRPC that are resistant to traditional chemotherapeutics and ARSIs. My laboratory is using transposon-mediated mutagenesis to guide the discovery of new druggable targets in drug-resistant CRPC. Transposon-mediated mutagenesis (i.e., Sleeping beauty 100X-SB100X) represents a power tool for inducing gain- and loss-of-function gene mutations that confer drug-resistance to ARSIs and taxane-derived chemotherapeutic drugs in cellular models of CRPC. Our discoveries will verify established mechanisms of drug-resistance in CRPC, but also uncover novel resistance mechanisms and pathways as druggable targets in the treatment of drug-resistant CRPC.
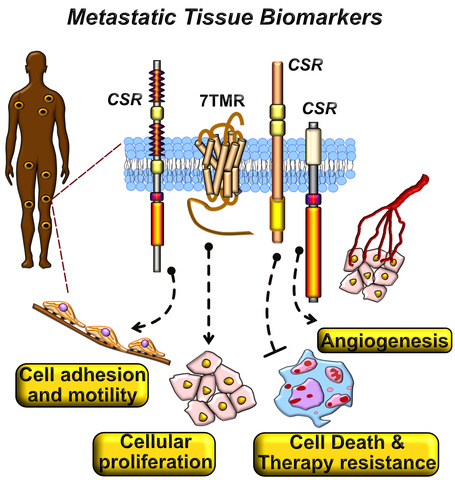
Biomarkers of Prostate-Derived Neoplasms
We are leveraging the power of quantitative mass spectrometry to define plasma membrane biomarkers in tissue samples of clinical prostate cancer. We are optimizing proteomic workflows and methods to quantify membrane-localized glycoproteins in fresh-frozen tissue samples. We are also interested is defining membrane glycoprotein expression signatures in patient-derived organoids of metastatic and castration-resistant prostate cancer. Our proteomic efforts seek to identify cell surface receptor (CSR ) expression changes underlying the pathobiology of metastatic and drug-resistant prostate cancers. These proteomic explorations will characterize seven transmembrane receptors (7TMRs) and CSRs as potential druggable targets in metastatic prostate cancers and provide new insights into the biology of functional interactions mediated by 7TMRs/CSRs in metastatic prostate cancers.
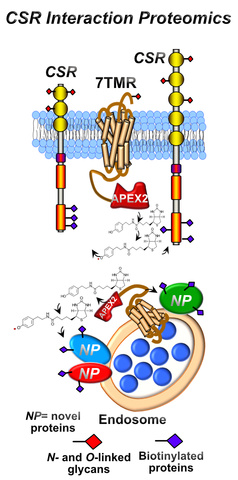
Cell Surface Receptor Interactomes
We are interested in mapping and defining protein interaction networks in 7TMRs and CSRs involved in prostate tumorigenesis. We have focused on chemokine receptor 7 (CXCR7) function in prostate-tumor cells because it is androgen-regulated and confers resistance to ARSIs. Moreover, CXCR7 is broadly expressed in multiple organ systems and CXCR7 functions have been implicated in multiple cellular processes such proliferation, differentiation, and cell survival. To better understand how CXCR7 engages these cellular processes at the molecular level, we are utilizing proximal labeling technology (i.e., APEX2) coupled to quantitative mass spectrometry to map the CXCR7-interactome in prostate-tumor cells. We want to understand how CXCR7 communicates with other CSRs at the plasma membrane to modulate CSR-dependent signaling responses in epithelial, skeletal muscle, and neurons. This proteomic investigation will provide new insights into CXCR7 functions at the molecular level and provide opportunities to modulate cellular processes through CXCR7-dependent signaling in homeostatic and pathological states.
Collaborations
We are actively seeking new collaborations with oncologists, urologists, and pathologists to profile prognostic and drug-resistant biomarkers in diseased tissue samples. This includes both epithelial neoplasms and hematologic malignancies. For further inquiries, please contact Dr. Wright at michael-e-wrightatuiowa.edu.